Proteomic Platform
Proteomics Platform
Welcome
At the Proteomics Platform we provide high quality standards to proteomic projects originated in Private companies, and Clinical or Academic entities. Since we are aware that proteomics is much more than just protein identification, we care for the development and settling of new proteomic technologies and methodologies.


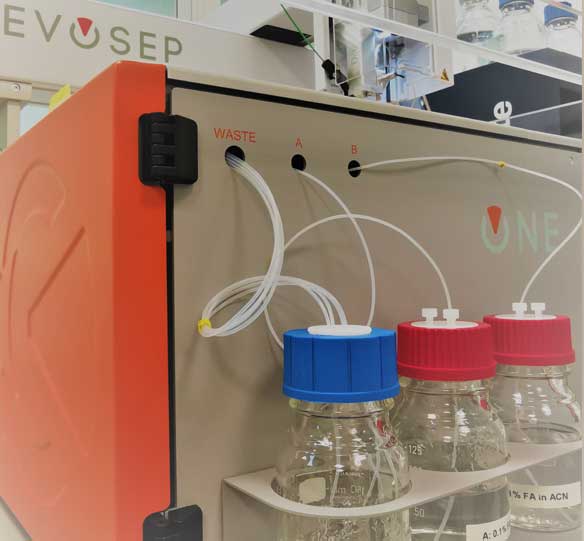
Proteomics through relative protein quantification has become a powerful tool to decipher the molecular basis of cellular processes and to identify potential biomarkers. With this in mind, we employ label-free quantification based on nLC-MS/MS technology. Our platform features one of the most advanced proteomic setups available: the EVOSEP ONE nanoLC system coupled online to a TIMS TOF HT mass spectrometer.
This cutting-edge combination delivers high sensitivity, performance, and throughput, enabling a robust proteomics pipeline. The result is a streamlined workflow that allows us to extract meaningful biological information from a wide variety of sample types—including tissue samples, biological fluids, total cell lysates, exosomes, viruses, and more. This versatility positions us to address challenges in translational and precision medicine.
- Platform Manager: Félix Elortza
Instrumentation
The proteomics platform is equipped with a wide range of analytical instrumentation and also state of the art mass spectrometers.
For protein characterization, identification and quantification, we have three mass spectrometers.
- UltrafleXtreme MALDI TOF (Bruker)
- EVOSEP ONE (and nano Elute, Bruker) coupled on-line to a TIMS Tof Pro mass spectrometer (Bruker).
EVOSEP ONE, a brand-new generation of nLC system, allows an unprecedented chromatographic speed, reproducibility and robustness.
Our tims TOF HT mass spectrometer is powered by PASEF (Parallel Accumulation–Serial Fragmentation) technology, which enables the alignment of precursor and fragment ion spectra based on ion mobility values when acquiring in data dependent acquisition mode (dda-PASEF). Moreover, we can apply dia-PASEF, a data independent acquisition (DIA) mode that uses PASEF technology to attain superior speed, comprehensive and ultra-sensitive quantitative proteomics.

Service catalogue
Our mass spectrometers are highly complementary and allow us to accomplish a wide range of proteomic analyses and services:
- Peptide Separation.
- Protein and Peptide molecular weight analysis determination by MALDI-TOF.
- Protein identification by nLC MS/MS.
- Protein relative quantification by label free nLC MS/MS.
- Protein Characterization: Post-translational Modification analysis *.
- Natural Peptidome (endogenous peptides) analysis by nLC MS/MS *.
*Contact Platform Manager for more information about these services. Please, do not hesitate contacting us if you would require a service which is not listed. We will study different possibilities on how to tackle the analysis..
Service Request
For information about experimental design and quote contact directly Felix Elortza (proteomicsplatform@cicbiogune.es).