2025/08/05
A proteomics study identifies remodeling of the liver ubiquitome and NEDDylome as a hallmark of Metabolic Dysfunction-Associated Steatotic Liver Disease (MASLD)
A new study led by researchers at CIC bioGUNE and IIS Biobizkaia, published in npj Gut and Liver (Nature Portfolio), uncovers a distinctive post-translational modification (PTM) signature involving ubiquitin and the ubiquitin-like molecule NEDD8 in Metabolic Dysfunction-Associated Steatotic Liver Disease (MASLD).
These findings deepen our understanding of the disease's pathophysiology and open new avenues for targeted therapeutic strategies.
Metabolic Dysfunction-Associated Steatotic Liver Disease (MASLD), previously known as non-alcoholic fatty liver disease (NAFLD), is nowadays the most common cause of chronic liver disease and the leading cause of liver-related morbidity and mortality worldwide. The environmental challenges of the liver lipid-enriched environment happening in MASLD are associated with a dynamic and rapid rearrangement of the liver proteome.
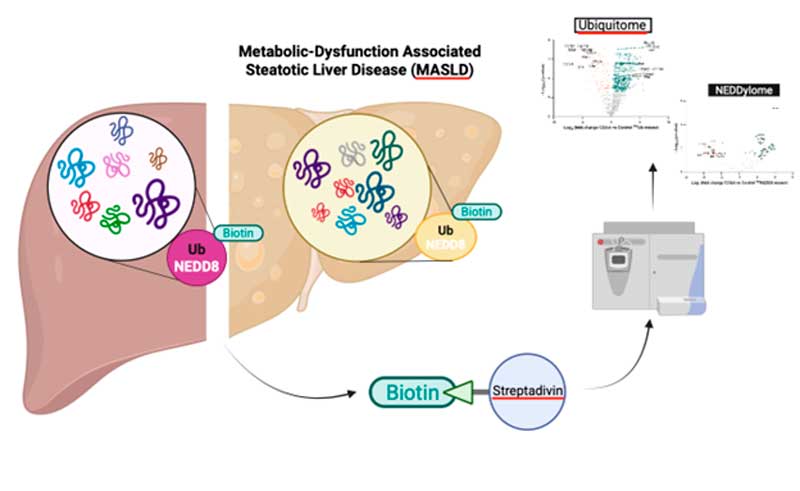
In this study, researchers have shown that these rapid responses of the liver proteome to environmental challenges observed in MASLD are associated with alterations in the patterns of protein post-translational modifications (PTMs), in particular modifications mediated by ubiquitin and the ubiquitin-like peptide, NEDD8.
Using advanced mass spectrometry-based proteomics and reporter transgenic mouse models, the team—working in collaboration with Ugo Mayor, the Proteomics Core Facility- SGIKER at EHU, and key contributors Rosa Barrio (principal investigator) and Félix Elortza (Proteomics Platform Manager) at CIC bioGUNE, member of BRTA, and CIBERehd —successfully identified the set of ubiquitinated and NEDDylated proteins that are differentially expressed in the liver of a diet-induced MASLD mouse model.
Quantitative mass spectrometry identified over 1,400 ubiquitinated proteins, with more than 600 proteins significantly altered in MASLD. Ubiquitinated proteins were enriched in processes related to endoplasmic reticulum and peroxisomal lipid metabolism, while NEDDylated targets primarily localized to mitochondrial pathways. Proteomic analysis revealed impaired proteasome function, endoplasmic reticulum stress, and mitochondrial dysfunction, suggesting that dysregulation of ubiquitin-like PTMs plays a critical role in MASLD.
“The identification of ubiquitin and NEDD8-modified proteins whose expression is altered specifically in MASLD is the first step to explore future MASLD therapeutic approaches based on targeting these proteins’ ubiquitin-like post-translational modifications. The profiling of hepatic ubiquitin-like PTMs is nowadays possible due to the advances of mass spectrometry technology combined with the utilization of reporter transgenic mouse models specifically designed to tag with biotin the proteins that are modified by the covalent binding of ubiquitin and NEDD8 peptides,” explain Drs. Malu Martínez-Chantar and Teresa Cardoso Delgado, researchers involved in the coordination of this work.
Photograph by: © Figure created using Biorender.
Reference: Teresa C. Delgado, Benoît Lectez, Marina Serrano-Maciá, Fernando Lopitz-Otsoa, Leire Uraga-Gracianteparaluceta, Ibon Martínez-Arranz, Jesús M. Arizmendi, Kerman Aloria, Mikel Azkargorta, Félix Elortza, James D. Sutherland, Rosa Barrio, Ugo Mayor & María Luz Martínez-Chantar. Rearrangement of the liver ubiquitome and NEDDylome in metabolic dysfunction-associated steatotic liver disease. npj Gut Liver. DOI: 10.1038/s44355-025-00032-0.
About CIC bioGUNE
The Centre for Cooperative Research in Biosciences (CIC bioGUNE), member of the Basque Research & Technology Alliance (BRTA), located in the Bizkaia Technology Park, is a biomedical research organisation conducting cutting-edge research at the interface between structural, molecular and cell biology, with a particular focus on generating knowledge on the molecular bases of disease, for use in the development of new diagnostic methods and advanced therapies.
About BRTA
BRTA is an alliance of 4 collaborative research centres (CIC bioGUNE, CIC nanoGUNE, CIC biomaGUNE y CIC energiGUNE) and 13 technology centres (Azterlan, Azti, Ceit, Cidetec, Gaiker, Ideko, Ikerlan, Leartiker, Lortek, Neiker, Tecnalia, Tekniker y Vicomtech) with the main objective of developing advanced technological solutions for the Basque corporate fabric.
With the support of the Basque Government, the SPRI Group and the Provincial Councils of the three territories, the alliance seeks to promote collaboration between the research centres, strengthen the conditions to generate and transfer knowledge to companies, contributing to their competitiveness and outspreading the Basque scientific-technological capacity abroad.
BRTA has a workforce of 3,500 professionals, executes 22% of the Basque Country's R&D investment, registers an annual turnover of more than 300 million euros and generates 100 European and international patents per year.
See a large version of the first picture