2012/06/27
Viruses find new ancestors
- Biogune researchers and scientists from the Universities of Oxford and Helsinki propose a radically different structure-based classification of viruses.
- Virus classification has been traditionally based on their genetic material and on the host cells they infect.
- This new organization shows significant similarities between viruses that infect very different organisms such as bacteria and humans.
We are surrounded by viruses. And we are not referring to those that "infect" our computers. We are referring to those microscopic infectious agents that multiply in the cells of other organisms, like plants, bacteria, archaea and animals, including human beings.
As an example, when we swim in the sea we are surrounded by millions of these microscopic agents (e.g. there are ~107 viruses per millilitre only on the sea surface). We only know an infinitesimal number of these infectious agents, and, of this small group, the scientific community has only managed to decode the 3-D structure of a limited number of them.
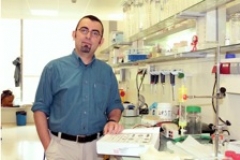
Researchers at the Structural Biology Unit at the Biogune, in collaboration with scientists of the Universities of Oxford and Helsinki, have challenged the usual classification of these agents provoking a true "conceptual revolution", in the words of Nicola G.A. Abrescia, Ikerbasque Professor at Biogune and one of the researchers participating in the review published in the prestigious Annual Review of Biochemistry, one of the scientific journals with the highest impact factors.
Viruses are basically composed of a genome (DNA or RNA) and a capsid, in other words, a protein structure that envelopes and protects this genetic material. Some have a lipid vesicle enveloping the capsid and, in other cases, the vesicle is surrounded by the capsid. Until now, viruses were classified considering their genetic material or the type of "hosts" that they infected.
This classification is broadly accepted by the international scientific community. However, this classification has certain inconveniences: it is complex, it does not classify all the known viruses and it does not establish any sort of relation between viruses infecting different types of organisms.
In this study they have shown that some viruses have capsids with very similar proteins, even though they infect completely different organisms. Traditional classifications consider that these viruses have no relationship between themselves, because they do not infect the same type of organisms.
Review
This scientific review has been authored by Prof. Nicola Abrescia, and worldwide recognized scientists involved in the study of viruses such as Prof. Dennis Bamford, of the University of Helsinki, Dr. Jonathan Grimes and Prof. David Stuart, of the University of Oxford and the latter also Director of Life Sciences of the Diamond Synchrotron in the UK.
This scientific team has presented a comprehensive analysis of the data accumulated over the last two decades and have proposed a new way of classifying the viral universe, which is radically different from what has been used until now.
"The publication of this review in this scientific journal represents a recognition of our proposal, since it shows the impact and relevance of this new way of classifying viruses", says Abrescia on the impact of the article by being published in such an internationally renowned journal.
New proposal
In this new proposal they have not considered which organism the viruses infects but its identity, considering as identity (or virus 'self') the 3-D structure of the protein forming the capsid. Logically, this scientific breakthrough can only focus on the small range of viruses "decoded" by the scientific community, i.e. those viruses in which the structure of the protein of the capsid has been identified.>
Of the 82 families of viruses that had been proposed in a previous classification (2008), the researchers have managed to group almost half in four large families, which implies an important simplification of the classification of the virus world by establishing (until now) the four main families mentioned above.
"The virus taxonomy as it is today is sometimes hard to grasp, we have tried to rationalise it. Apparently unrelated viruses, are now related with our new approach. By using the capsid structure we have generated four viral lineages, which simplifies the vision of the virus world", say Abrescia.
Additionally, the system proposed by the researchers in the Annual Review of Biochemistry allows to relate viruses that were considered completely different since they infect different organisms.
For example, they have noted that, in terms of the structure of the capsid, adenoviruses that infect humans causing respiratory diseases are similar to PRD1, which infects bacteria. With this approach it has been possible to detect a partial similarity between the capsid of herpes virus and phage HK97.
These findings, highlights Abrescia, "could open the door to the use of similar strategies against viruses that had not been considered related so far but that share similar capsid structures."
Ancestral viruses
Another implication deriving from the new classification is the fact that the viruses affecting different domains of life (bacteria, animals, etc.) are so similar, thus indicating a common ancestor origin, which would be prior to the separation in the three domains of life millions of years ago.
In other words, it is believed that all living creatures descend from one shared universal ancestor (LUCA). This organism evolved in different ways, thus originating the current domains (archaea, bacteria y eukaryote).
The fact that some viruses infecting very different domains, such as humans and bacteria, share very similar capsid protein structures, could imply that they come from shared ancestral viral lineages prior to the separation of the living creatures in the aforementioned branches.
Nicola G.A. Abrescia is Ikerbasque Professor hosted by the CIC bioGUNE at the Structural Biology Unit. Abrescia's research is focused on studying very large macromolecular complex such as viruses, with particular emphasis to those viruses with a lipidic vesicle such as Hepatitis C virus and bacteriophage PRD1, using electron microscopy and X-ray crystallography techniques.
References for the study
Structure Unifies the Viral Universe.
Nicola G.A. Abrescia, Dennis H. Bamford, Jonathan M. Grimes, and David I. Stuart. Annual Review of Biochemistry, July 2012.
http://www.annualreviews.org/doi/abs/10.1146/annurev-biochem-060910-095130
See a large version of the first picture