2015/01/26
Structural Basis of Ligand Binding to UDP-Galactopyranose Mutase fromMycobacterium tuberculosis
Mycobacterium tuberculosis is a human pathogen that causes tuberculosis, a disease that kills ca. 2 million people worldwide every year. The occurrence of multidrug and extensively drug-resistant strains of M. tuberculosis have emerged in a way that they are resistant to most or all known antibiotics. Thus, there is an urgent need to develop new drugs against tuberculosis.
Galactofuranose (Galf) is a key component of the polysaccharide that connects the peptidoglycan and the mycolic acid layers in mycobacteria cell wall. One key enzyme involved in Galf metabolism is UDP-Galactopyranose mutase (UGM). This enzyme is essential for mycobacterial growth. Since Galf and UGM are not found in humans, UGM is a validated therapeutic target.
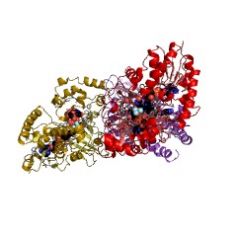
Within an international collaboration with research groups at Canada (Sanders), United Kingdom (Linclau) and Belgium (Vincent), the group of J. Jiménez-Barbero at the Infectious Disease Programme at CIC bioGUNE has contributed to understand the molecular basis of the mechanism of action of UGM, which catalyzes the reversible conversion of UDP-galactopyranose (UDP-Galp) to UDP-galactofuranose (UDP-Galf). The structure of different complexes formed by UGM with polyfluorinated glycomimetic inhibitors has been determined, showing the structural and dynamic features of the molecular recognition process and opening the gate to the development of drugs able to target this enzyme. They have demonstrated that the enzyme selects a high-energy conformation of the furanose within the catalytic pocket. This fact has important implications from the structure-based-drug-design point of view. From a basic research perspective, this research also has shown the huge perspective for employment polyfluorination for the design of novel glycan-based mimetics.
Link to the article here
See a large version of the first picture