2014/03/13
S-adenosylmethionine levels regulate the Schwann cell DNA methylome
Axonal myelination is essential for rapid saltatory impulse conduction in the nervous system, and malformation or destruction of myelin sheaths leads to motor and sensory disabilities. Schwann cell myelination is a highly orchestrated and complex developmental process, which requires coordinated and widespread changes in gene expression. The establishment and maintenance of this gene expression pattern is principally regulated by an elaborate network of transcription factors. Recently, several studies have shown that another level of transcriptional control is provided by recruitment of the NuRD and BAF chromatin-remodeling complexes to regulatory regions of key myelin genes, and possibly by histone deacetylation by HDAC1 and HDAC2. DNA methylation is an essential epigenetic modification during mammalian development, yet its role in myelination remains obscure.
In this study, led by Dr Ashwin Woodhoo from the Metabolomics Unit at CIC bioGUNE, in collaboration with scientists from the CIBERehd, Instituto Universitario de Oncología del Principado de Asturias (IUOPA-HUCA), Bellvitge Biomedical Research Institute (IDIBELL), University College London and University of Southern California, the role of DNA methylation in controlling gene expression during Schwann cell myelination and in the development of diabetic neuropathies was examined. Using high-resolution genomic maps to examine the methylome dynamics during Schwann cell myelination in vivo, the authors found that Schwann cell myelination was characterized by global DNA demethylation, which was associated with activation of critical myelination-associated genes, particularly those involved in synthesis of lipid, a critical component of myelin. Importantly, their observations support the notion that levels of SAMe play a critical role in regulating the methylation status of myelinating Schwann cells. Peripheral nerves from mice with enforced elevated levels of SAMe were characterized by DNA hypermethylation globally, and at promoter and enhancer regions of several lipid synthesis genes. Furthermore, a link was found between reduced SAMe levels and DNA demethylation in peripheral nerves of a mouse model of diabetic neuropathy, suggesting a possible role of SAMe in the pathogenesis of this disease.
Taken together, this study shows that Schwann cell myelination is characterized by widespread DNA methylation changes that likely play a key gene regulatory role. These methylation dynamics are dependent on a tight control of SAMe levels, and imbalances to its levels likely lead to aberrant DNA methylation patterns and consequently gene expression changes during development of neuropathies.
This study was recently published in the journal Neuron.
Reference to the article:
Marta Varela-Rey*, Marta Iruarrizaga-Lejarreta*, Juan José Lozano*, Ana María Aransay*, Agustín F. Fernandez, José Luis Lavin, David Mósen-Ansorena, María Berdasco, Marc Turmaine, Zigmund Luka, Conrad Wagner, Shelly C. Lu, Manel Esteller, Rhona Mirsky, Kristján R. Jessen, Mario F. Fraga, María L. Martínez-Chantar, José M. Mato, Ashwin Woodhoo. S-adenosylmethionine Levels Regulate the Schwann Cell DNA Methylome (2014). Neuron 81(5): 1024-1039. * first authors
LINK
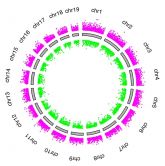
Figure Legend:
Chromosome ideogram showing distribution of hypomethylated (green) and hypermethylated (red) differentially methylated tiling regions in SAMe-treated Schwann cell cultures
See a large version of the first picture