Activity Detail
Seminar
Diverse mass spectrometry applications for the analysis of proteomes, proteins, and peptides.
Felix Elortza, PhD
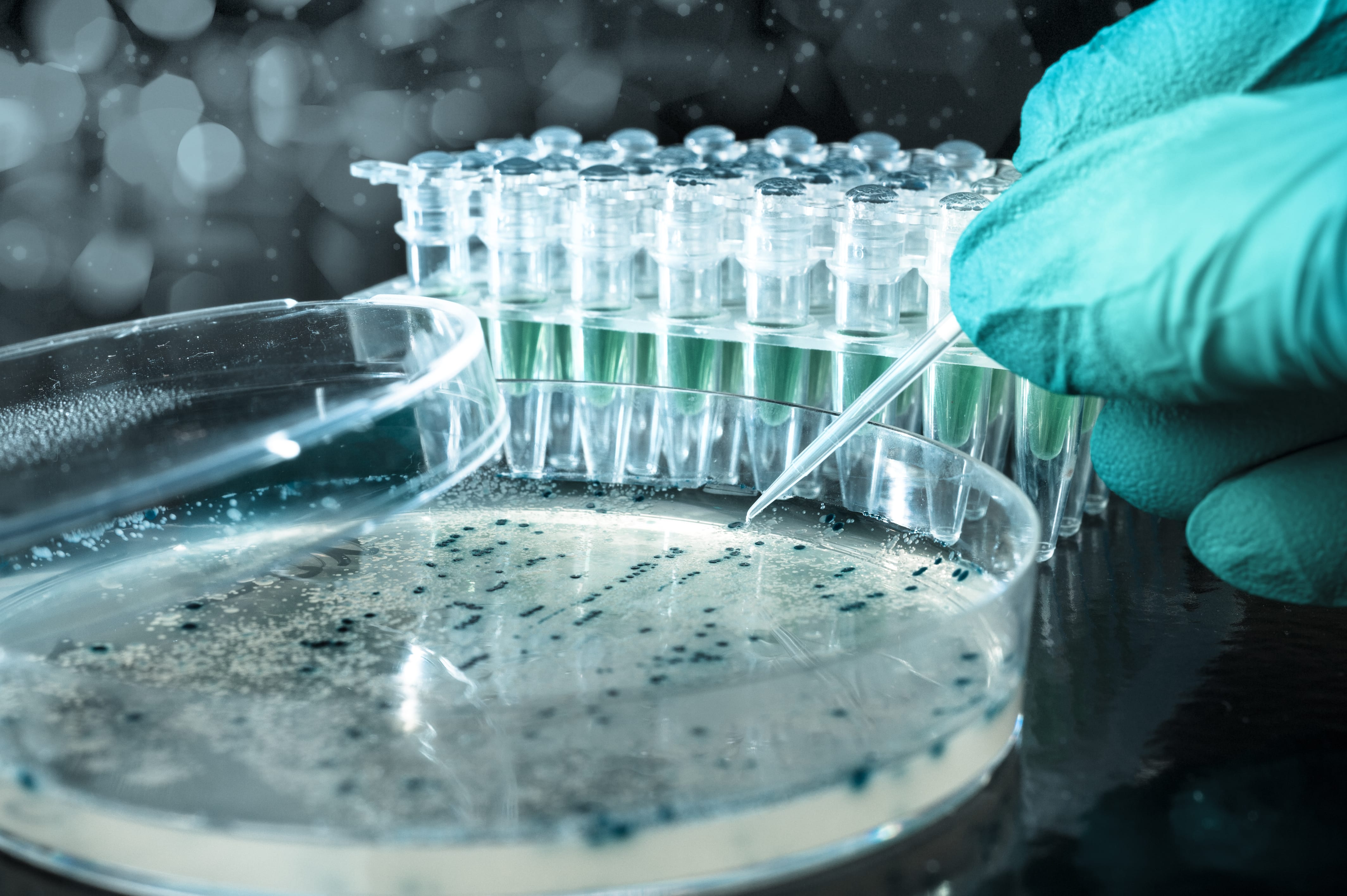 In less than two decades, proteomics has evolved from cumbersome 2D gels and purified single-protein analysis using peptide mass fingerprinting via MALDI-TOF mass spectrometry, to the identification and quantification of hundreds/thousands of proteins by nanoscale liquid chromatography coupled on-line to tandem mass spectrometry (nLC MS/MS). This particular configuration, which is much faster, sensitive and suitable for automatization has allowed proteomics to be applied to projects with scarce material, such as those using clinical samples. Moreover, the combination of nLC MS/MS with specific sample preparation methodologies allows in-depth protein analyses such as exploring post-translational modifications.
Proteomics a priori is focused on protein analysis, although most current analytical approximations are performed on peptides derived from proteins after a specific protease digestion (e.g trypsin). The study of endogenous short peptides is an emerging and exciting trend; for instance, a peptide derived from a precursor protein may display a different activity than the precursor itself, and peptides encoded by small unannotated ORFs may have important functions. This new field of peptidomics can provide complementary information to other “-omics” about ongoing biological processes.
The Proteomics Platform fosters the analysis of proteins and peptides by mass spectrometry. We have optimized a diverse catalog of methods to cover a wide range of analyses. In this seminar, a survey of different Proteomic applications from our lab will be shown, such as Label Free Relative Protein Quantification, MALDI-TOF Tissue Imaging MS, and Endogenous Peptidomic Analysis. Results obtained from different collaborations will be highlighted.
In less than two decades, proteomics has evolved from cumbersome 2D gels and purified single-protein analysis using peptide mass fingerprinting via MALDI-TOF mass spectrometry, to the identification and quantification of hundreds/thousands of proteins by nanoscale liquid chromatography coupled on-line to tandem mass spectrometry (nLC MS/MS). This particular configuration, which is much faster, sensitive and suitable for automatization has allowed proteomics to be applied to projects with scarce material, such as those using clinical samples. Moreover, the combination of nLC MS/MS with specific sample preparation methodologies allows in-depth protein analyses such as exploring post-translational modifications.
Proteomics a priori is focused on protein analysis, although most current analytical approximations are performed on peptides derived from proteins after a specific protease digestion (e.g trypsin). The study of endogenous short peptides is an emerging and exciting trend; for instance, a peptide derived from a precursor protein may display a different activity than the precursor itself, and peptides encoded by small unannotated ORFs may have important functions. This new field of peptidomics can provide complementary information to other “-omics” about ongoing biological processes.
The Proteomics Platform fosters the analysis of proteins and peptides by mass spectrometry. We have optimized a diverse catalog of methods to cover a wide range of analyses. In this seminar, a survey of different Proteomic applications from our lab will be shown, such as Label Free Relative Protein Quantification, MALDI-TOF Tissue Imaging MS, and Endogenous Peptidomic Analysis. Results obtained from different collaborations will be highlighted.